# | cache: true
# load necessary packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.preprocessing import StandardScaler
import repsim
# set a random seed
np.random.seed(1)
# prepare the prototype
iris = load_iris(as_frame=True).frame.iloc[:, :4]
url = "https://vincentarelbundock.github.io/Rdatasets/csv/datasets/USArrests.csv"
usarrests = pd.read_csv(url, index_col=0)
X = StandardScaler().fit_transform(iris.sample(50, random_state=1))
Y = StandardScaler().fit_transform(usarrests)
n, p_X, p_Y = X.shape[0], X.shape[1], Y.shape[1]
# generate 10 of each by perturbation
mats = []
for _ in range(10):
mats.append(X + np.random.normal(scale=1.0, size=(n, p_X)))
for _ in range(10):
mats.append(Y + np.random.normal(scale=1.0, size=(n, p_Y)))
# compute similarities
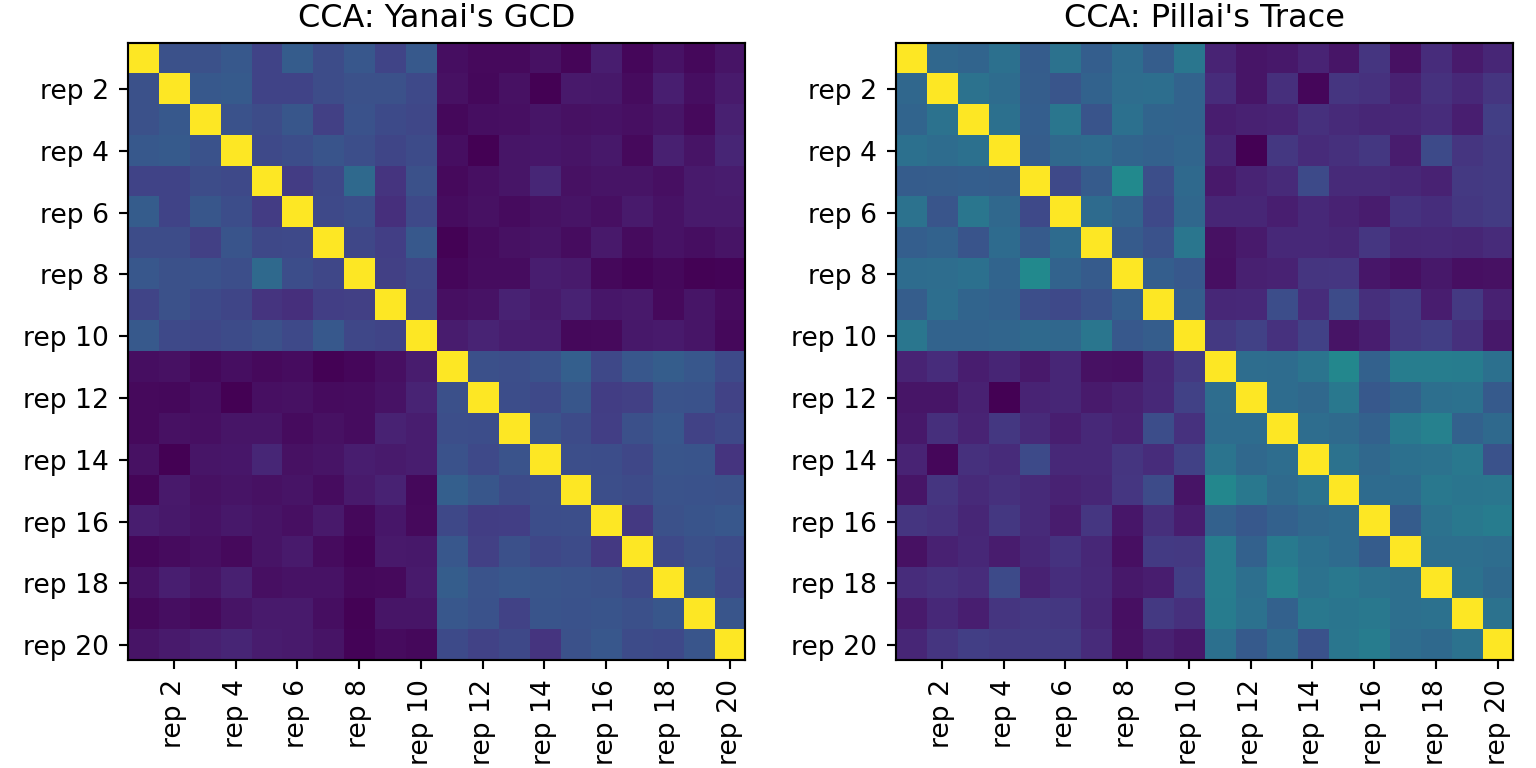
cca_gcd = repsim.cca(mats, summary_type="yanai")
cca_trace = repsim.cca(mats, summary_type="pillai")
# visualize: two heatmaps side by side
fig, axes = plt.subplots(1, 2, figsize=(8, 4), constrained_layout=True)
titles = ["CCA: Yanai's GCD", "CCA: Pillai's Trace"]
mats_show = [cca_gcd, cca_trace]
labs = [f"rep {i}" for i in range(1, 21)]
even_idx = list(range(1, 20, 2))
for ax, mat, title in zip(axes, mats_show, titles):
im = ax.imshow(mat, origin="upper")
ax.set_title(title)
_ = ax.set_xticks(even_idx)
_ = ax.set_xticklabels([labs[i] for i in even_idx], rotation=90)
_ = ax.set_yticks(even_idx)
_ = ax.set_yticklabels([labs[i] for i in even_idx])
plt.show()